Modeling and analysis of synthetic, methylation based, epigenetic gene circuits
Epigenetics are the study of changes to gene activity without altering the DNA sequence. In recent years it has gotten more attention from researchers as it plays a vital role in gene transcription and therefore many processes in the cell. It’s contribution in cancer and disease is also becoming more and more apparent. Apart from the role of epigenetics in in vivo functioning it can also be a very nice tool in synthetic biology.
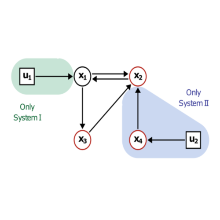
In this project our collaboration partners in the group of Albert Jeltsch, Department of Biochemistry, Institute of Biochemistry and Technical Biochemistry, have used DNA methylation, an important epigenetic mark, to create synthetic gene circuits. These circuits are designed to have two stable states, an off-state and an on-state. The transition between these can be triggered by different constructs. This also allows for a multitude of different trigger scenarios, like heat, presence of different metabolites, as well as DNA damage by radiation or DNA damaging agents.
On the mathematical modeling side of this project we try to replicate the dynamics of this system through a set of differential equations. We firstly calibrate the model to the experimental data using parameter optimization and employ methods to analyze the obtained model. Through this analysis we gain insight into the underlying biochemical model. Bifurcation analysis, for example, can aid in understanding the role of different parameters on the stable states of the system. With this information we can make recommendations for improvement of specific properties of the biochemical system to for example make the on- and off-state more stable or the system more or less sensitive to the trigger input.
Moreover we use a population model to reflect the diversity of the biological system. With this modeling approach we are able to distinguish different subpopulations, cells that stay reliably in the ON-state and cells that switch back to the OFF-state after differing duration in the ON-state. We will further make use of this population approach and expand it to fully use the newly available single cell measurements and through this be even more capable to reflect the heterogeneity in the cell population.
Overall this projects aims at a better understanding of the biological mechanisms involved and providing valuable input to further improve this synthetic system.

Nicole Radde
Prof. Dr. rer. nat.Professor for Mathematical Modeling and Simulation of Cellular Systems

Viviane Klingel
M.Sc.[Image: Ludmilla Parsyak, © Fraunhofer IAO]